I am interested in studying fluid flow and wall mechanics in interconnected networks of blood vessels. To date, I have worked on several interdisciplinary projects involving data analysis, modeling, and model analysis to predict pulmonary hemodynamics under normal and hypertensive conditions. Methodologies developed for these projects are applicable to study a wide range of problems in cardiovascular physiology.
Main Collaborators: Mette Olufsen, Mansoor Haider, Dirk Husmeier, Naomi Chesler
Broad Areas of Interest and Related Topics
Mathematical Modeling: Mathematical models of physiological systems are emerging as useful tools for exploring the underlying mechanisms of observed physiological phenomena. Significant advances in medical and computational technologies have enabled us to assimilate big data with mechanistic models, realizing the prospect of using these models in clinical situations.
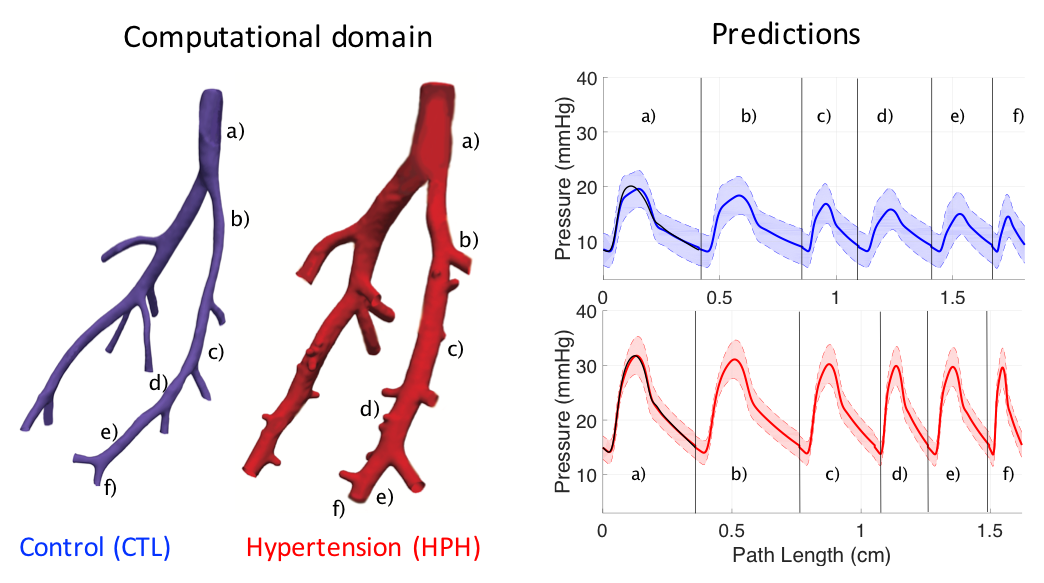
I develop 1D fluid dynamics models of vascular networks, derived form Navier-Stokes equations and a tube law describing elastic stress-strain relation for vessel wall tissues. The process includes the development of component models, their integration into a system level model, and numerical solution. These models are then used to simulate and explore disease etiologies, e.g., those associated with group-specific pulmonary hypertension, to understand how vascular remodeling (both tissue and network structure) affects the system level hemodynamics.This work involves research activities on several topic including:
- Arterial-venous coupling: fluid dynamics coupling of anatomical structures.
- Vascular wall mechanics: phenomenological and constitutive models describing linear or non-linear deformation of vessel wall.
- Microcirculation: Morphology based fluid dynamics or lumped (0D) parameter models of vascular beds.
- Ventricular-vascular coupling: coupling vascular networks with simplified models of pumping heart and dynamic valve states (in progress)
- Numerical analysis: Finite difference methods, numerical coupling or component models and boundary conditions
- Wave intensity analysis: decomposition of waveforms, wave reflections analysis
- Model validation: local and global validations, time and frequency domain validations
- Statistical model selection: assessing information gain versus computation cost of using various component models
Related publications
- MU Qureshi, MJ Colebank, LM Paun, L Ellwein, N Chesler, MA Haider, NA Hill, D Husmeier, MS Olufsen. Hemodynamic assessment of pulmonary hypertension in mice: a model based analysis of the disease mechanism, Biomech Model. Mechanobiol, In press, 2018
- MU Qureshi, MA Haider, NC Chesler, MS Olufsen, Simulating the effects of hypoxia on pulmonary hemodynamics in mice. In proceedings of the 5th Computational and Mathematical Biomedical Engineering, (CMBE17) pp. 271-274,2107, Pittsburgh, USA.
- PLee, BE Carlson, NC hesler, M Olufsen, MU Qureshi, NP Smith, T Sochi, DA Beard: Heterogeneous Mechanics of Mouse Pulmonary Arterial Network, Biomech Model Mechanobiol, 15(5): 1245 – 1261, 2016
- MU Qureshi, NA Hill, A Computational study of pressure wave reflections in the pulmonary arteries. J Math Biol 71(6): 1525 – 1549, 2015
- MU Qureshi, GDA Vaughan, C Sainsbury, M Johnson, CS Peskin, MS Olufsen, NA Hill, Numerical simulation of blood flow and pressure drop in the pulmonary arterial and venous circulation. Biomech Model Mechanobiol, 13(5): 1137 – 1154, 2014
- MU Qureshi, MA Haider, MS Olufsen, A numerical study of pulmonary vascular efficiency and right ventricular afterload during hypoxia induced pulmonary hypertension in mice. Accepted for presentation at 8th World Congress of Biomechanics, Dublin, Ireland, 8 – 12 July, 2018. (Peer reviewed abstract)
- MA Haider, MU Qureshi, NA Hill, MS Olufsen, Application of the HGO model tocapturing the pressure-area relationship in a large murine pulmonary artery under pulsatile flow. Accepted for poster presentation at 8th World Congress of Biomechanics, Dublin, Ireland, July 8 -12, 2018 (Peer reviewed abstract)
Parameter Estimation
Dealing with high dimensional parameter space is one of the major challenges in fluid dynamic network modeling. This aspect of my work focuses on structural and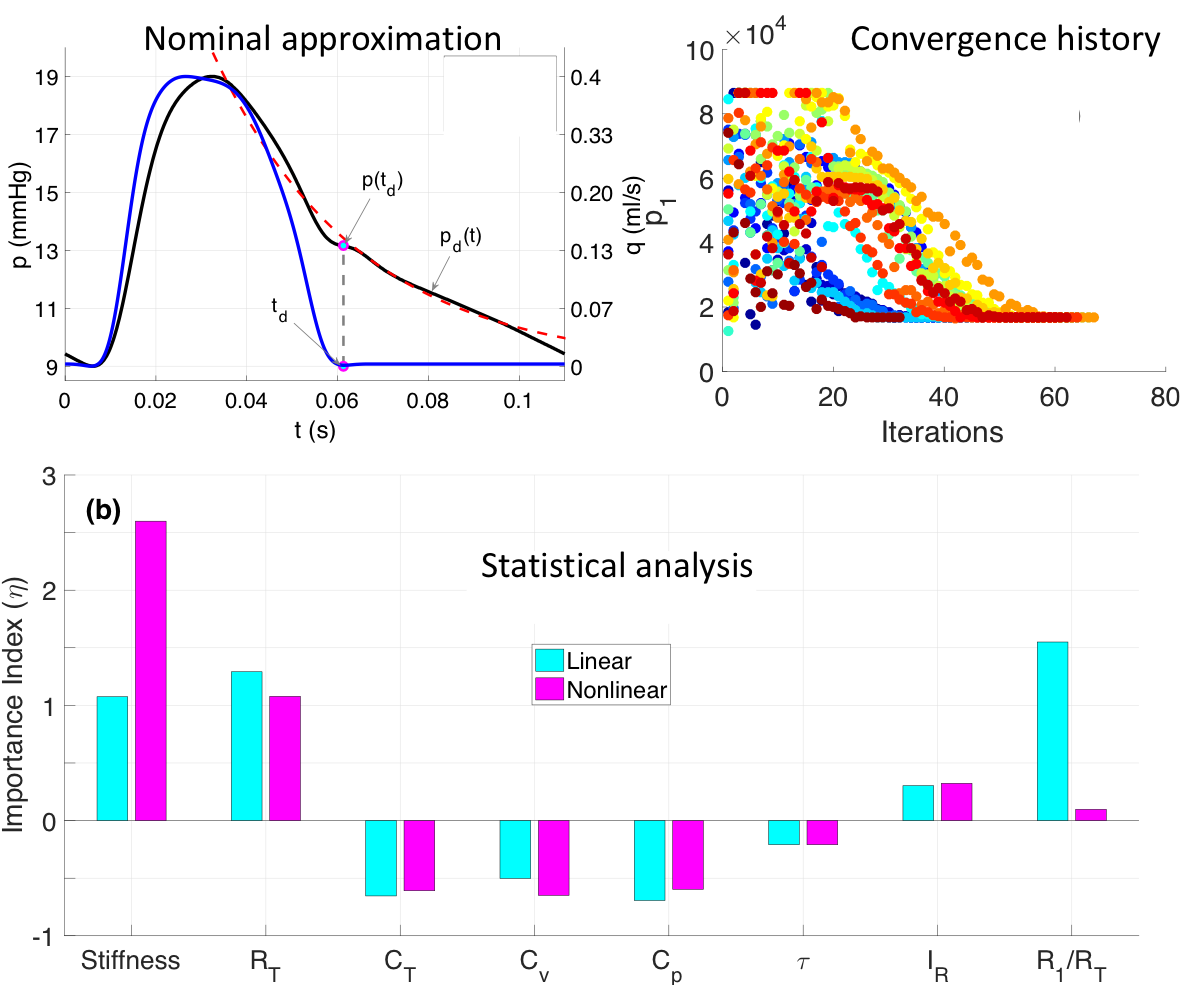 practical identifiability as well as estimation of physiologically relevant model parameters. My work on recent projects involve:
practical identifiability as well as estimation of physiologically relevant model parameters. My work on recent projects involve:
- Nominal parameter approximation: combining the prior knowledge (data and literature) and analytical results from simplified fluid mechanics problem (e.g. steady flow in an analogous network of rigid vessels) to compute nominal parameters.
- Sensitivity analysis: local and global sensitivity analysis
- Correlation analysis: analysis of covariance structure
- Optimization: constraint nonlinear optimization and its convergence
There are several open questions to this end of my work including analysis of temporal and spatial sensitivities of model parameters, reducing dimension via scaling relations, sensitivity to the quantity of interest.
Related publications:
- MU Qureshi, MJ Colebank, LM Paun, L Ellwein, N Chesler, MA Haider, NA Hill, D Husmeier, MS Olufsen. Hemodynamic assessment of pulmonary hypertension in mice: a model based analysis of the disease mechanism, Biomech Model. Mechanobiol, In press, 2018
- M Paun, MU Qureshi, MJ Colebank, NA Hill, MS Olufsen, MA Haider, D Husmeier, MCMC methods for inference in mathematical model of pulmonary circulation. Special Issue Article, Statistica Neerlandica, 72(3):306 – 338, 2018
- M Paun, MU Qureshi, MJ Colebank, MA Haider, MS Olufsen, NA Hill, D Husmeier, Parameter Inference in the Pulmonary Blood Circulation of Mice, In proceedings of the 32nd International Workshop on Statistical Modelling (IWSM 2017), pp. 190-195, 2017, Groningen, Netherlands
Uncertainty Quantification
One of the objectives of my research is to be able to predict patient-specific hemodynamics by using non-invasive data (obtained through MRI and CT scans). However, there are errors involved in every step of data handling, which propagate to the model predictions, affecting both parameters and the quantity of interest. This necessitates the need to propagate uncertainties, not only related to data (statistical uncertainty) but also due to modeling assumptions (scientific uncertainty), through the modeling process and quantify their effects on the model responses. My recent work combines data analysis with Bayesian and frequentist approaches to quantify uncertainties in the form of posterior parameter distributions and confidence and prediction intervals for the quantity of interest.
Related publications:
- MJ Colebank, MU Qureshi, MS Olufsen, Sensitivity analysis and uncertainty quantification for a 1D fluids model of pulmonary circulation (2018) Under review for publication. (preprint arXiv:1807.02863).
- Mitchel J. Colebank, M. Umar Qureshi, Naomi Chesler, Dirk Husmeier, Mette S. Olufsen, Laura M. Ellwein, Influence of image segmentation on fluid model predictions in the pulmonary circulation (Almost ready for submission in 2018).
- MJ Colebank, MU Qureshi, D Husmeier, NC Chesler, MS Olufsen, Uncertainty in 1D fluid dynamics network of pulmonary vasculature. Accepted for poster presentation at 8th World Congress of Biomechanics, Dublin, Ireland, July 8 -12, 2018 (Peer reviewed abstract)
- C Battista, A Arnold, MU Qurehi, MS Olufsen. Estimating boundary conditions for one-dimensional modeling of blood flow and pressure in arterial networks. In proceedings of the 4th International Conference on Computational & Mathematical Biomedical Engineering (CMBE15), pp 307-310, 2015, Cachan (Paris), France
Clinical Data Analysis
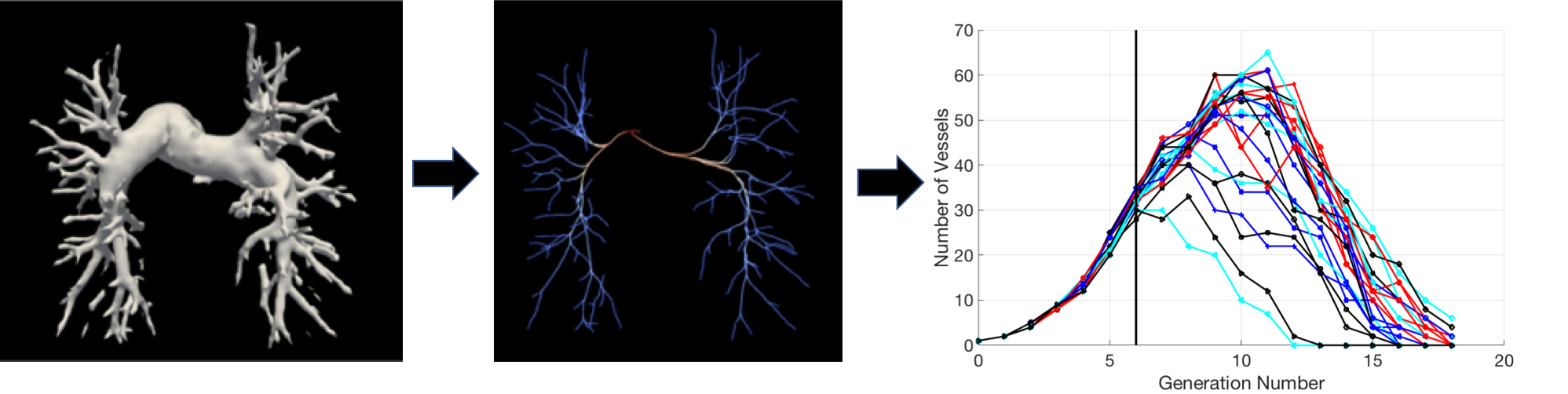
I am interested in clinical data analysis, not only because it informs and validates the modeling process, but also because it is extremely useful in identifying a disease marker. 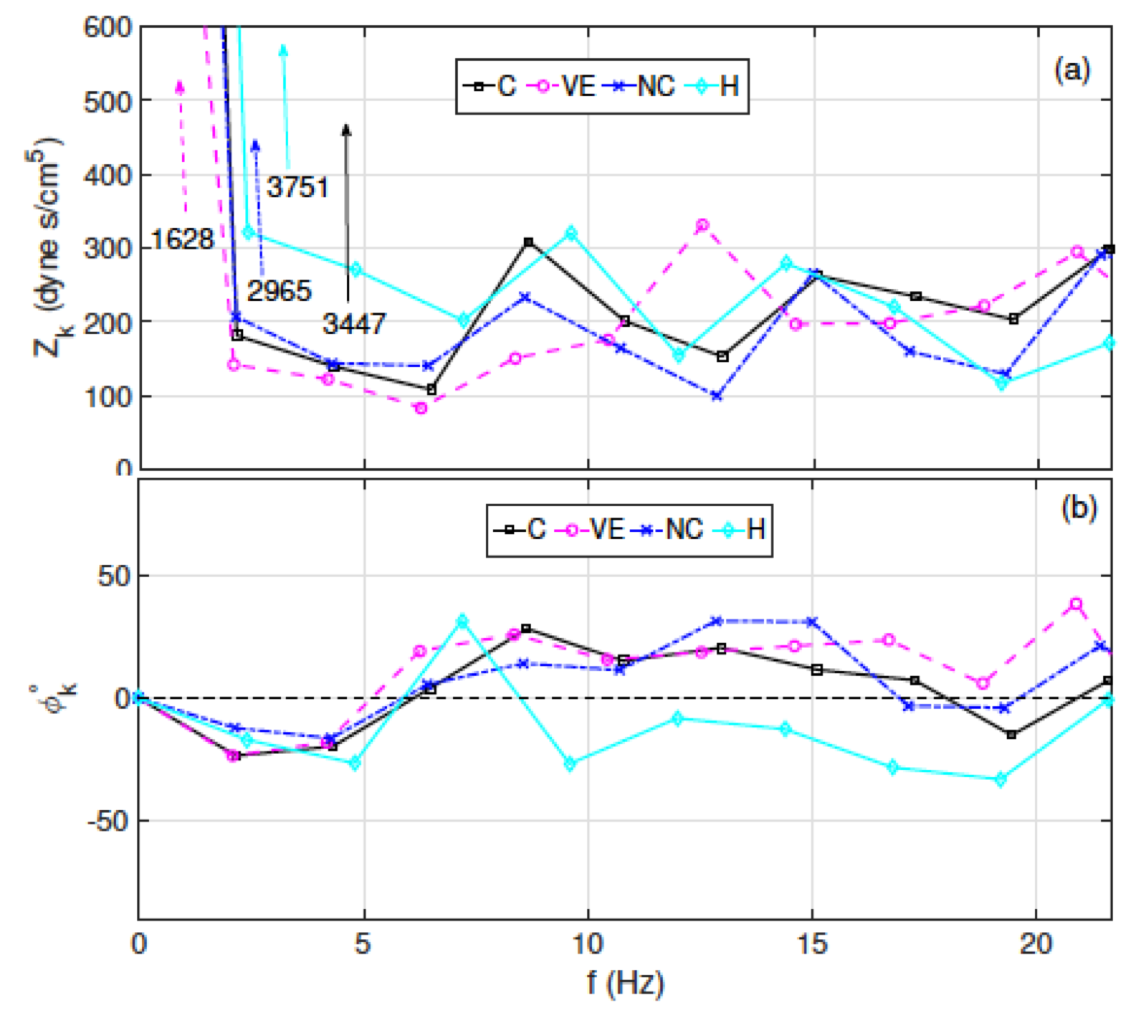 Specifically, I am interesting in developing/applying methods to visualize hemodynamic and morphology data with an aim to identify disease related abnormality at an early stage. In the future, I would like to explore the prospect of combining machine learning tools with theoretical approaches describing the tree networks (not specific to physiology). My current and prospective work on this topic includes:
Specifically, I am interesting in developing/applying methods to visualize hemodynamic and morphology data with an aim to identify disease related abnormality at an early stage. In the future, I would like to explore the prospect of combining machine learning tools with theoretical approaches describing the tree networks (not specific to physiology). My current and prospective work on this topic includes:
- Hemodynamic signal analysis: in time (wave intensity analysis) and frequency (impedance analysis) domains
- Medical image analysis: applications of image segmentation algorithms, 3D to graph structure, analysis of fractal dimension, topological data analysis, big network visualization
Clinical Applications
One of the applications of my models includes numerical assessments of techniques, used to approximate quantities pertinent to a disease prognosis. My work on this aspect includes
- Numerical assessment of clinical metrics: used model and data analysis to study methods of estimation for pulse wave velocity and characteristic impedance. Both are metrics of arterial stiffness, which is an independent predictor of mortality in patients with pulmonary hypertension.
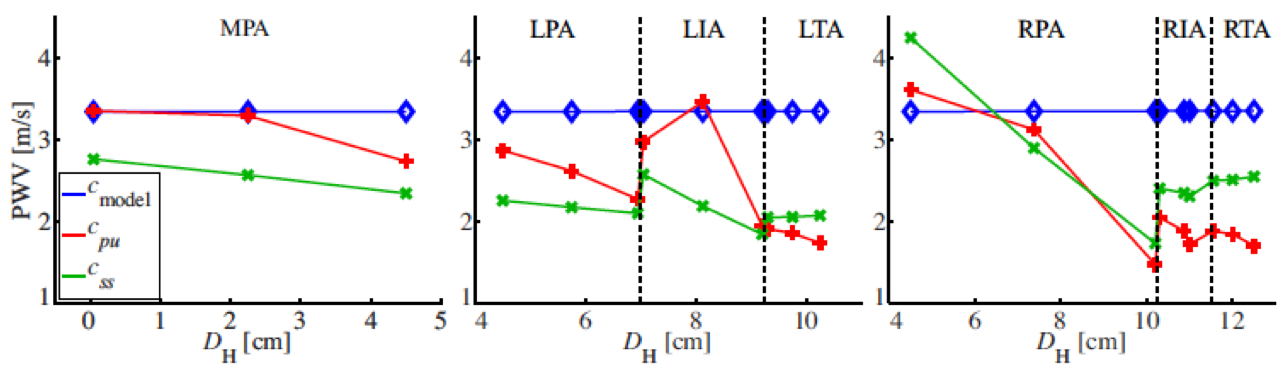
Related publications
- MU Qureshi, MJ Colebank, D Schreier, DM. Tabima, MA Haider, NC Chesler and MS Olufsen, Characteristic Impedance: Frequency or time domain approach?, Physiol Meas. 39(1):014004, 2018
- MU Qureshi, NA Hill, A Computational study of pressure wave reflections in the pulmonary arteries. J Math Biol 71(6): 1525 – 1549, 2015
